Assay for Inhibitors of Nonsense-Mediated RNA Decay
Simpler, faster, and less expensive than exogenous NMD assays Ease of use in vivo or in tissue and organs without the need to transfect reporter genes into cells.
Use of endogenous NMD assay to screen for compounds and genes that modulate PTC and/or NMD
None
20180010188
Background An estimated 9 million people in the US suffer from genetic disorders that are a consequence of a nonsense or frameshift mutation that introduces a premature termination codon (PTC). PTC is also known as a nonsense codon or premature stop codon and such codons lead to loss of protein products. Nonsense-mediated RNA decay (NMD) is a cellular quality control mechanism to selectively degrade transcripts that have a PTC codon. Currently exogenous assays are used to screen for compounds that inhibit NMD.These labor-intensive exogenous assays use PTC-containing reporter genes and are restricted to certain transfectable cells. Since these reporter genes may not transfect the entire cell population in an assay, exogenous assays have limited sensitivity. Exogenous reporters also have a high false positive rate, because these assays are simultaneously affected by non-NMD regulation. Thus new and effective assays to screen for NMD inhibitors are desirable. Images Fig. 1 shows the schematics of the novel endogenous assay that uses RT-qPCR methods to detect NMD and non-NMD isoform of the same gene. The NMD isoform can be the inclusion or the exclusion isoforms. The inclusion isoform is detected by primer F1 and isoform specific primer R1. The exclusion isoform is detected by F2 and isoform-specific junction primer R2. Primers F3 and R3 are commonly used in alternative splicing assays to detect both inclusion and exclusion isoforms. Related Materials Tech ID/UC Case 27517/2016-708-0 Related Cases 2016-708-0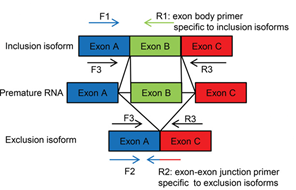
USA


